We are thrilled to announce an exciting evolution in our hackathon day, a cornerstone of the SWAT4HCLS event. Traditionally, our participants have gathered on the last day in an unconference format to delve into hacking, discussions, and drafting new project proposals and research papers. Building on last year’s collaboration with the renowned BioHackathon – an event with its roots in Japan and now celebrated globally, including significant editions like the Elixir BioHackathon – we are deepening our alignment.
Our audience has consistently shown interest in SWAT4HCLS conferences and various biohackathons worldwide. This shared enthusiasm has been evident in recent research papers where authors have credited both SWAT4HCLS and the Biohackathon(s). Acknowledging this and reflecting the spirit of biohackathons, we transitioned last year to naming our hackathon day as Biohackathon-SWAT4HCLS.
This year, we are taking another exciting step forward. While the last day remains dedicated to the hackathon, we will extend the Biohackathon-SWAT4HCLS experience throughout the event. By offering a dedicated biohackathon space on the preceding days, participants can delve deeper into their hack projects, culminating in the presentations that traditionally close the SWAT4HCLS event. Join us in this enriched experience and dive deeper into innovation and collaboration!
Call for Project Proposals for Biohackathon-SWAT4HCLS!
As we embark on this extended journey of the Biohackathon-SWAT4HCLS, we invite all enthusiastic researchers, developers, and innovators to submit their project proposals. This is a golden opportunity to bring your ideas to the forefront, collaborate with like-minded professionals, and work intensively to bring them to life during the biohackathon.
Whether you have a developing concept that needs refining, a challenge that requires collective brainstorming, or a mature idea ready for hands-on development, we want to hear from you. By submitting your proposal, you can harness the collective expertise and passion of the SWAT4HCLS community.
Submission Guidelines:
- Provide a clear title for your project.
- Offer a concise description of the problem or challenge you aim to address.
- Outline the goals and expected outcomes of the project.
- Mention any specific tools, technologies, or datasets you plan to use.
- If possible, specify the ideal team size and any required expertise for collaborators.
The proposals will be featured during the biohackathon, providing teams ample time and space to work on them, leading to the grand showcase on the last day of SWAT4HCLS.
Don’t miss out on this chance to shape the future of bioinformatics, healthcare research, agricultural research, biodiversity research and their integration with the semantic web. Submit your project proposal now and be a part of the groundbreaking Biohackathon-SWAT4HCLS!
Submit your proposal here.

Biohackathon Pitches
1. Publishing FAIR Datasets from Bioimaging Repositories
Project lead: Josh Moore (German Bioimaging)
Project Description
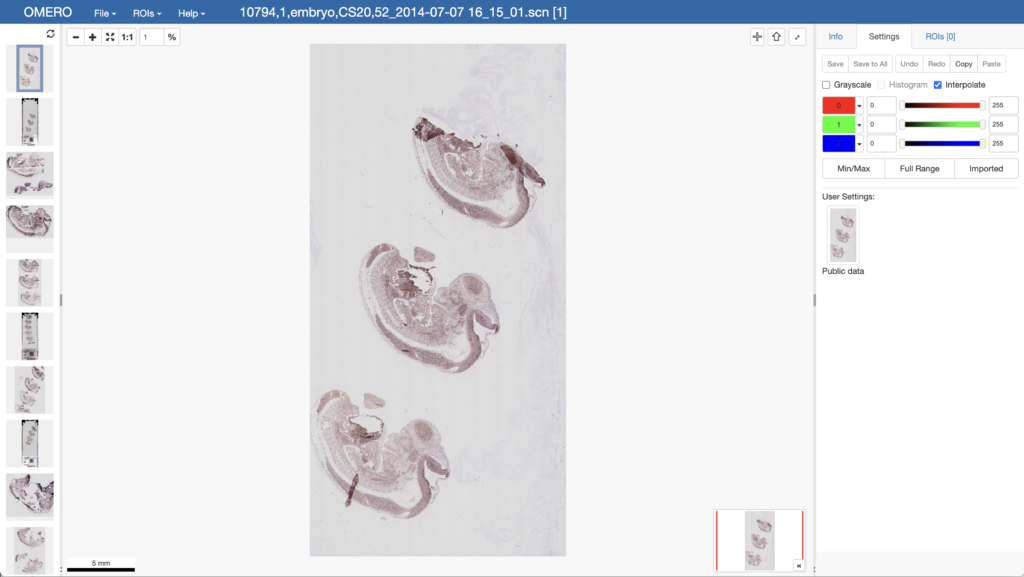
The Image Data Resource (IDR) is home to 13 million multi-dimensional image datasets. Each of these is annotated with (a subset of) Gene, Phenotype, Organism/Cell Line, Antibody, siRNA, and Chemical Compound metadata. This data is stored in a data management system named OMERO, where it is stored in PostgreSQL tables.
Initial work has been performed to export this information as RDF from OMERO using https://pypi.org/project/omero-rdf (See the related SWAT4HCLS Poster)
The export of the largest single study (defined as a collection of the image datasets associated with a single publication), however, generates 100M triples. This study representing images of tissue from the Human Protein Atlas has been exported directly using SQL and parallelized scripts.
Continuing work from last year’s hackathon this year, we would like to:
- Capture the output from bioimaging repositories that use OMERO like IDR and SSBD in GitHub
- Publish this representation to Zenodo to acquire a DOI
- Use this backend for hosting a Fair Data Point (FDP)
Expertise Needed
Familiarity with RDF (incl. but not limited to SPARQL and ingestion/query optimization) is required.
Familiarity with Fair Data Points or more generally DCAT would be beneficial.
2. Exploring a full semantic representation for OMOP
Project lead: Andrea Splendiani
OMOP is a widely used standard for Real World Evidence, that is, the description of patients (or people) health histories for population level studies. While OMOP present a rich approach to normalise terminologies for values, it falls short of a full semantic representation, as the main model is semantic: classes and properties don’t have a formal representation, neither their are defined and related to the broader context of healthcare and life sciences ontologies.
Objective of this project is to explore a full semantic representation of OMOP, and perhaps try a few queries that cross between OMOP data and “-omics” resources.
Expertise needed
Knowledge of OMOP, knowledge of semantic technologies, basic scripting (or proficiency with a specialised RDF-ization tool), some knowledge of SPARQL.
3. Derive a drawing standard inspired by UML, ER, dot, Cytoscape, to graphically describe data shapes as Shape Expressions
Project lead: Andra Waagmeester
Shape Expressions (ShEx) provides a formal (machine actionable) language to describe linked-data schemas. Ways to draw those data shapes do exist, but are primarily done in loosely structured manner. It is still needed to manually craft those boxologies into ShEx.
In this hackathon topic we would like to identify a notation standard using graphical depictions of existing data shape templates. We can take inspiration from either UML or ER diagrams.
Ideally the resulting graphical diagrams depicting the linked-data shapes can be automatically stored as shape expressions, preferably directly, otherwise to bidirectional conversion tool
The ultimate goal is to have the means to draw shape expressions, while initially not having to go through the steep learning curve that comes with ShEx.
Expertise needed
People knowledgable on underlying graphical notation formats (UML, ER, dot, graphviz, cytoscape, etc).
4. AI Prompt engineering for SPARQL, ontologies, etc
Project lead: Andra Waagmeester
This is a proposal for designating a AI prompt engineering corner where we can explore various strategies to engineer a prompt to get the expected the results. A github repository will be created for this Biohackathon topic where we will store expectation, necessary prompts, their results and possible conclusions on the accuracy of this.
Creating prompts to get effective SPARQL queries is the first topic that comes to mind, but I am also thinking to explore prompts to align biological databases with linked data repositories by aligning with the linked-data repositories community standards ((bio)schema.org, wikidata, obo, snomed, FHIR etc).
Expertise needed
AI, SPARQL, OWL, JSON, CSV, API’s
5. Linked-data games corner
Project lead: SWAT4HCLS
This is a proposal to set up a games corner, where participant to SWAT4HCLS could explore and play existing board/computer games with a life science theme. Playing those games alone is fine to relax after a intense SWAT4HCLS day, but ultimatly we would like to see some new games emerging from the linked-data we are exploring during SWAT4HCLS.
6. Implementing ISA mapping with DCAT
Project lead: Liao, XiaoFeng
A FAIR Data Point (FDP) is a software architecture aiming to define a common approach to publish semantically-rich and machine-actionable metadata according to the FAIR principles (findable, accessible, interoperable and reusable). FDP ultimately stores information about data sets, which is the definition of metadata. The FDP uses the W3C’s Data Catalog Vocabulaire (DCAT) version 2 model as the basis for its metadata content.
In X-omics initiative, considering the various metadata formats adopted by the different X-omics communities, it is reasonable to adopt a standard metadata format as a template for submitting the metadata. To this purpose, we employed the ISA metadata framework as our basic framework to capture and standardize study (design) information from the different -omics metadata schemes. The ISA metadata schema is commonly adopted by the research community for submission of metabolomics data, for example by EMBL-EBI’s MetaboLights.
Objective:To make ISA metadata schema compatible with FDP, we need to align the ISA metadata schema with DCAT.Specifically, to implement the mapping of Investigation/Study/Assay with DCAT core
7. Exploring how FAIR data resources on rare diseases can benefit people living with a rare disease
Project lead: Marco Roos
Do you wish to contribute to the quality of life of people living with a rare disease by showing how semantic data can be exploited?
There are thousands of rare diseases, many start at childhood, and the effects are often severe and life threatening. Each disease has sparse data scattered around the world. Therefore, the European Joint Programme Rare Diseases produced guidelines for data resources that are relevant for rare diseases to become part of a FAIR principles based ‘Virtual Platform’.
The ‘machine actionable’ requirement of the FAIR principles is addressed by ontological models for resource descriptions (based on the Data Catalog Vocabulary), and an ontological model ‘Care-SM’ for data elements inside the data resources. These are served via ‘FAIR Data Points’ (REST calls into the semantic layer), and tools were built to help with data conversions or mapping to other popular models or APIs. As more resources connect to the network, an increasingly large virtual large knowledge graph of ‘ontologised’ data and metadata will emerge. We wish to demonstrate what benefits this will bring for patients. For the conference we created a small test bed environment to allow participants of SWAT4HCLS to be creative with the ontology-based semantic layer of this emerging VP network.
We hope to seed ideas for innovative applications for the benefit of rare disease patients.
Expertise needed
SPARQL
Working with knowledge graphs
AI on semantic data
8. Beyond the VoID
Project lead: Ana Claudia Sima
Upset about slow federated queries? Frustrated about the lack of visualization tools that give you insights into what a knowledge graph actually contains? Come hack with us for improvements to the current community practice!
We use and provide extended VoID descriptions in our SPARQL endpoints, and would like it if you would too!
We have a tool called VoID generator that generates detailed descriptions for RDF datasets loaded into named graphs. It works for large knowledge graphs too – UniProt included!
We use these descriptions for many purposes – to generate knowledge graph schema diagrams, to improve autocomplete for SPARQL queries, as well as linking documentation to example queries. We’re open to new ideas on how to adapt them for new purposes!
Do you have a dataset and would like to generate a shape file, for further documentation and / or validation? Or in general have an interest in having better (visual) documentation of a SPARQL endpoint? Join us at the Biohackathon!
Expertise Needed
None, but please bring some data 😉 along with an open mind, willingness to try out some code and enthusiasm to bring a contribution towards going beyond the VoID in knowledge graph documentation!
9. Guidelines for data captures systems to consume ontologies and controlled vocabularies
Project lead: Simon Jupp
Organisations are increasingly working to standardised ontologies and terminologies to increase the interoperability of data as part of their FAIR data strategy. To ensure data is “Born FAIR” there is a need for the systems (ELNs, LIMS etc) that capture the data to surface these terminologies to users at authoring time. Whilst these systems may support controlled terminologies, they are often closed systems and integrate poorly with centralised terminology services. This leads to data management systems becoming siloed and hampers data interoperability across systems.
As large organisation looks to centralise the management of terminology assets to ensure data is being generated to a set of common standards, there’s a need to support vendors of data management system to utlise these terminologies to maximise the investment going into standard terminology development. We are looking to form a group of interested parties to develop a set of guidelines that can be followed for programmatic integration of terminology services into third-party software. The guidelines will aim to minimise the effort for systems to query and lookup terminology from a terminology server. These guidelines are built up from a core set of common use-cases and can be built upon over time to deal with more elaborate use-cases.
An example of a such a guideline would be to establish a common mode of interaction between two systems for looking up a standard term by a preferred or alternate label.
For example, a terminology service should provide the following functionality:
- A stateless (REST) API endpoint that takes two parameters as input. One parameter is a query string to be lookup up by the terminology service, the second optional parameter is the name or identifier of the terminology being looked up.
- Authentication to the API endpoint can be optional and should be provided using an encrypted API token in the request header
- The terminology service should respond with an array of standards terms in a common format (JSON). Each entry for a term in the response should provide at minimum the identifier for the term and a preferred label. Additional, optional fields in the response could include a term definition and weighting for the result.
We can already see in practice that some popular terminology services already meet these requirements, despite having slightly different API contracts. Two public ontology services, NCBI BioPortal and EBI’s Ontology Lookup Service, and SciBite’s CENtree Ontology Management System, all provide APIs that meet these requirements for terminology lookup.
Despite the widespread availability of ontology services and standards like RDF and SPARQL, large software vendors have been slow to adopt these standards and often resort to simpler mechanisms to consume and utilise ontologies and CVs. This creates a barrier for the adoption of ontologies and exacerbates the challenges around data harmonisation and interoperability.
Expertise Needed
- Ontology/terminology Service Providers
- Expertise in RDF, OWL, SPARQL and REST APIs
- Experience working with popular ELN or LIMS systems
10. Semantic Phenopackets for federated learning/analytics
GA4GH Phenopackets is a standard to represent computable clinical data of patients and integrate it with genomic data. While Phenopackets is appropriate to represent patient level data, it is not clear how to link this data to omics resources that aggregate patient information for population studies. In order to address this challenge, we started using semantic phenopackets to represent biosample metadata for individuals who participated in genotype-phenotype studies and publish it from a FAIR Data Point. To follow-up the BioHackathon Europe 2021 project on the modelling of semantic Phenopackets for the Semantic Web and FAIR federated learning/analytics, we propose enhancing interoperability between metadata description of omics analyses and metadata of individual patient data for federated learning/analysis using Phenopackets, FAIR and semantic technologies. Our objective during the BioHackathon is to investigate and harmonise metadata described in other federated ecosystems, and maybe perform queries crossing omics population level studies with phenopackets individual data.
Expertise Needed
- GA4GH Phenopackets
- RDF, OWL and SPARQL
- knowledge representation, metadata modelling
- FAIR Data Points and GA4GH Beacon API
